Data Distillation in Medical Imaging
Reducing Data Dependency · Part 3
Medical imaging faces persistent data challenges: strict privacy constraints, high annotation costs, limited and imbalanced datasets, and the logistical burden of storage and inter-site data transfer. These issues were discussed in detail in the Data Bottleneck article.
As the demand for robust and clinically trustworthy AI grows, so does the interest in techniques that reduce dependence on large labelled datasets while maintaining strong performance. In the Reducing Data Dependency series, previous articles explored data-efficient learning paradigms such as semi-supervised, unsupervised and self-supervised learning, as well as augmentation and synthetic data generation.
Data distillation is a natural continuation of these concepts.
Why Data Distillation in Medical Imaging
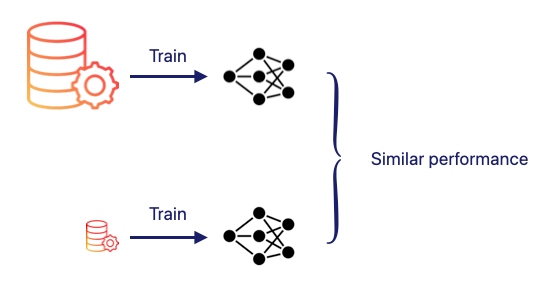
Efficient inter-site data sharing is becoming increasingly important, yet the sensitive nature of medical imaging demands strict privacy, security and regulatory safeguards. Data distillation offers a practical way to capture the essential information contained in a dataset without transferring the original data. It compresses a large dataset into a much smaller, distilled version that enables efficient training and deployment while preserving comparable model performance. Crucially, the distilled dataset can often be exchanged more easily in settings where sharing raw patient data is restricted.
However, it remains unclear whether techniques that succeed on natural images transfer directly to medical imaging. Medical images present unique challenges: Class variability is often lower, anatomical structures are visually similar, and clinically meaningful distinctions rely on extremely subtle changes in texture, density or morphology. Small patches from radiology or pathology images may appear nearly identical, and distinguishing benign from malignant lesions – or different disease stages – requires expert interpretation of fine-grained visual cues.
From my experience auditing datasets and supporting AI development teams, most failures do not come from the distillation algorithm itself but from upstream data issues: artefacts, scanner drift, unbalanced cohorts, among others. Distillation can compress a dataset, but it cannot fix weak data foundations. When applied on well-curated datasets, however, it becomes a powerful tool for reducing storage, compute and cross-site sharing constraints.
This article reviews two approaches that are particularly relevant for the field: Dataset Condensation (DC) and Matching Training Trajectories (MTT).
Dataset Condensation (DC)
Dataset Condensation compresses a large training dataset into a much smaller synthetic dataset, sometimes as few as 10 to 50 images per class. The goal is to preserve the key statistical and structural information needed for model learning so that a model trained on the distilled data can reach performance close to one trained on the full dataset.
In other words:
Think of it as compressing an entire dataset into a handful of highly informative synthetic examples.
These synthetic samples are not simply selected from the original dataset. Their pixel values are optimised so that training a model on them reproduces the learning signal generated by the real dataset. In practice, the pixels of the synthetic images are treated as trainable parameters that are updated to reproduce how the model would behave if trained on the original dataset.
Data Condensation with Curriculum Gradient Matching
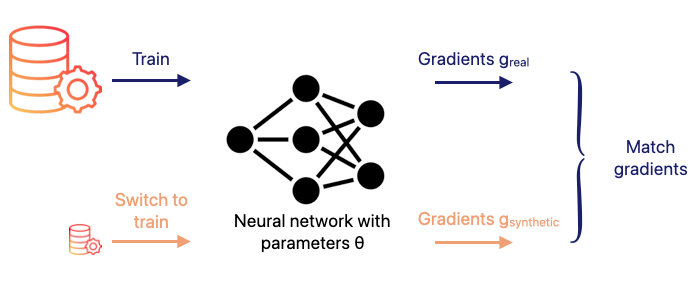
The objective is to generate a synthetic dataset S such that a model trained on S learns in a similar way to a model trained on the full dataset T. This is done by ensuring that, at each optimisation step, the gradients produced by the synthetic data resemble those produced by the real data.
A neural network with parameters θ is used to compute:
- the gradient of the loss on a minibatch of real images
- the gradient of the loss on a minibatch of synthetic images
The synthetic data S is then updated so that these two gradients become as close as possible. In essence, the dataset condensation reduces to matching the gradients of both loss functions, allowing the synthetic dataset to encode the same learning signal as the original data.
Distillation by Matching Training Trajectories (MTT)
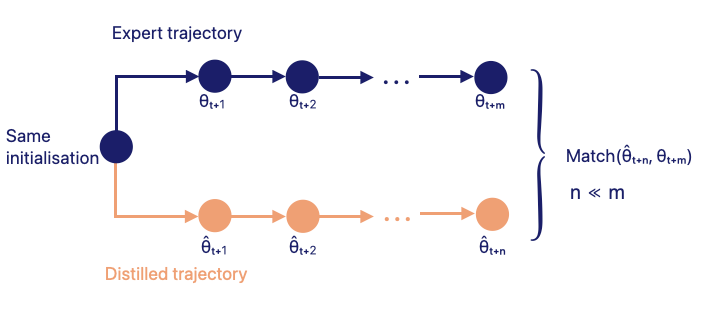
Matching Training Trajectories (MTT) creates synthetic data that guide a model along a sequence of weight updates resembling the sequence obtained when training on real data from the same initialisation.
MTT performs long-range parameter matching. After N updates on synthetic data, the model parameters should be close to those obtained after M updates on the real dataset, with M greater than N in practice. By bringing these two sets of parameters closer, the distilled dataset gradually learns to encode the key training signal of the full dataset.
How does this work
- The student model is initialised with parameters taken from a random point in the real-data training trajectory.
- The student model is trained for N gradient descent steps using the current synthetic dataset.
- From the real or expert trajectory, parameters M steps ahead of the sampled starting point are retrieved. These represent how the model evolves when trained on real data.
- A weight-matching loss is computed between the student’s parameters after N updates and the expert’s parameters after M updates.
- The synthetic images are updated until the distilled dataset reliably guides the student model along trajectories that resemble those of the real dataset.
Benefits
- Faster and lower cost training
- Smaller datasets to store, handle and share
- More compliant workflows, since raw patient data does not need to be exchanged
Application
Li et al. evaluated Dataset Condensation (DC) and Matching Training Trajectories (MTT) on eight datasets from the MedMNIST collection : PathMNIST, DermaMNIST, OCTMNIST, BloodMNIST, TissueMNIST, OrganAMNIST, OrganCMNIST and OrganSMNIST.
Across these benchmarks, DC outperformed both MTT and random selection.
Distillation methods showed clear advantages on several datasets such as BloodMNIST, TissueMNIST, OrganAMNIST, OrganCMNIST and OrganSMNIST, while offering limited gains on others. This indicates that distillation effectiveness is task dependent.
Li et al. also reported a strong linear correlation between the accuracies of distilled subsets and randomly selected subsets. This suggests that random selection acts as a proxy for task difficulty. Simpler classification tasks with more separable features are more likely to benefit from distillation, while complex tasks may require tailored or domain-aware approaches.
Conclusion
Data distillation reduces data requirements by creating compact synthetic datasets that preserve key learning dynamics. Approaches like DC and MTT show clear potential in medical imaging, especially on tasks with distinct and easily separable features. Yet results vary across datasets, suggesting that distillation is not a one-size-fits-all solution and must be adapted carefully to each use case.
As part of a broader data-efficient strategy, it remains a valuable tool for building scalable, privacy-aligned medical AI systems while helping teams reduce their dependency on large, fully supervised datasets.
From my experience building and auditing medical imaging pipelines, distillation should be viewed as a complementary tool rather than a shortcut. Where it shines is reducing compute, enabling privacy-preserving collaboration, and supporting early prototyping, not replacing real clinical data. When integrated thoughtfully, it can significantly reduce reliance on large datasets while maintaining clinical credibility.