Training Models With Little or No Labels
Reducing Data Dependency · Part 1
After discussing the data bottleneck problem in medical imaging in the previous article, this new series explores practical ways to reduce data dependency and ultimately overcome the data bottleneck.
Although the examples are drawn from medical imaging, the bottleneck, and the strategies to mitigate it, are central to many other computer vision fields as well.
Introduction
In the previous article, we explored why data scarcity remains one of the biggest challenges in medical AI. Labeled data is expensive, inconsistent, and slow to produce – on top of all the burdens related to transfer, storage, curation, annotation, and regulatory constraints.
There are, however, many ways to address this data bottleneck – from generating synthetic data to distilling large models or automating annotations.
Before diving into these approaches, let’s start with one powerful family of solutions: learning with little or no labels.
Three major learning paradigms have emerged to overcome the lack of annotated data: unsupervised, semi-supervised, and self-supervised learning, each approaching the problem from a distinct angle.
With recent breakthroughs, models are increasingly able to learn effectively without relying on large labeled datasets.
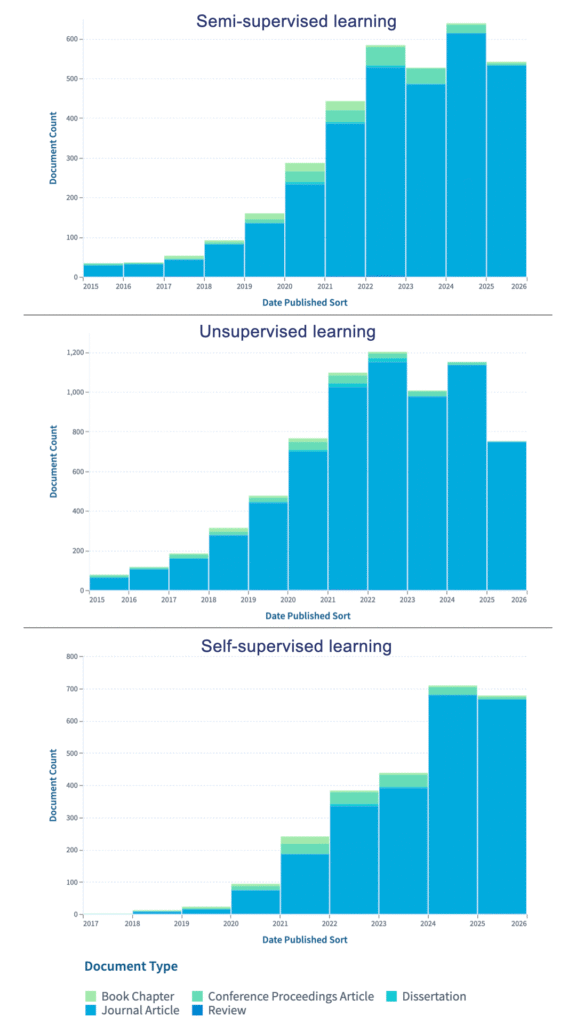
This trend is mirrored in the research landscape: publication counts show a sharp rise in work on semi-supervised, unsupervised, and self-supervised learning since 2020, reflecting the field’s rapid shift toward methods that reduce reliance on labeled data.
In this article, we’ll introduce these three paradigms at a high level. In the following articles of this series, we’ll explore each one in greater detail.
Semi-supervised Learning
Semi-supervised learning combines labeled and unlabeled data within the same training process. It leverages a small set of labeled samples together with a much larger pool of unlabeled ones .
Deep semi-supervised learning benefits from the strong feature extraction capabilities of deep neural networks while using unlabeled data to improve robustness and generalisation . The goal is to achieve supervised-level performance while relying on fewer true labels.
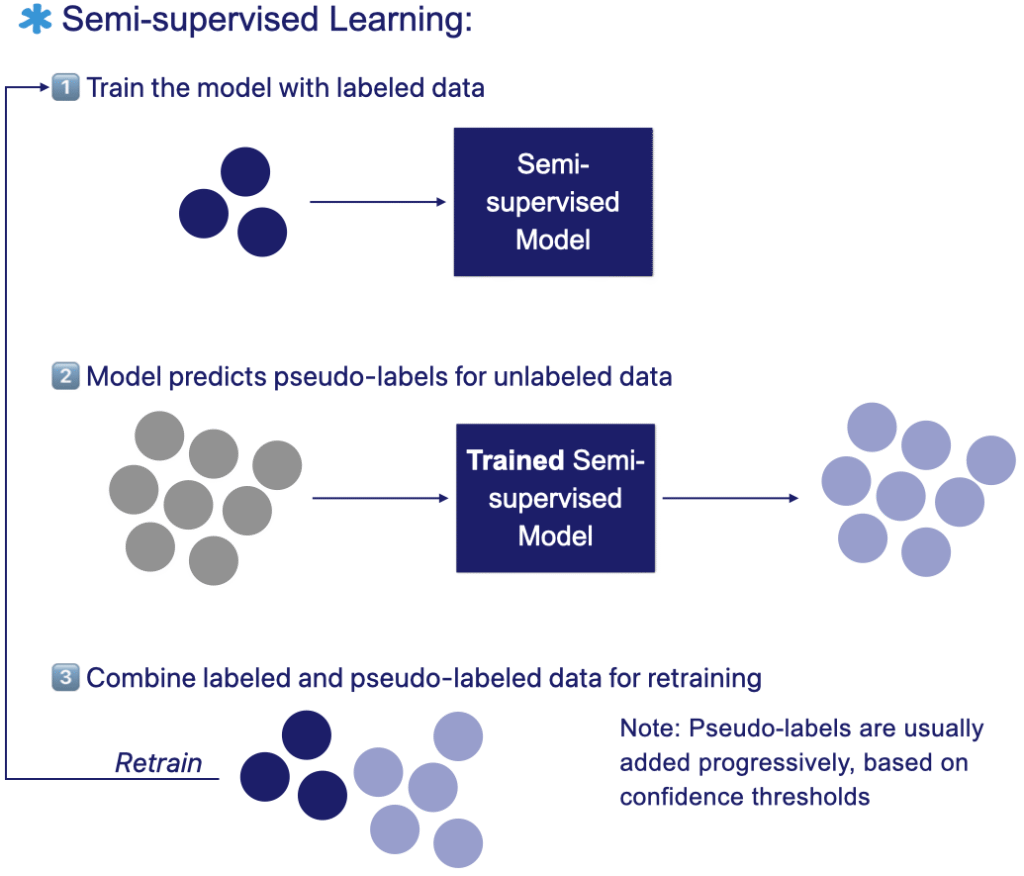
A simple and widely used strategy is pseudo-labeling. In this approach, a model first trained on the labeled subset generates temporary labels for the unlabeled data based on its own predictions. It is then retrained using these pseudo-labels, effectively expanding the training set without additional manual annotation. This practical method allows the model to exploit far more data than was originally labeled.
Applications in medical imaging include:
- Classification: Huynh et al. introduced a new loss function to handle class imbalance in semi-supervised medical image classification. Their Adaptive Blended Consistency Loss (ABCL) improved performance on skin cancer and glaucoma fundus datasets.
- Segmentation: Rieu et al. proposed a semi-supervised framework for brain segmentation in FLAIR MRI.
- 3D Detection: Wang et al. developed FocalMix: a semi-supervised method for 3D medical image detection, evaluated on CT datasets for lung nodule identification.
The figure below illustrates the semi-supervised learning workflow: the model is first trained on a small labeled dataset. It generates pseudo-labels for the unlabeled samples then retrains iteratively on both labeled and unlabeled datasets to improve performance.
Unsupervised Learning
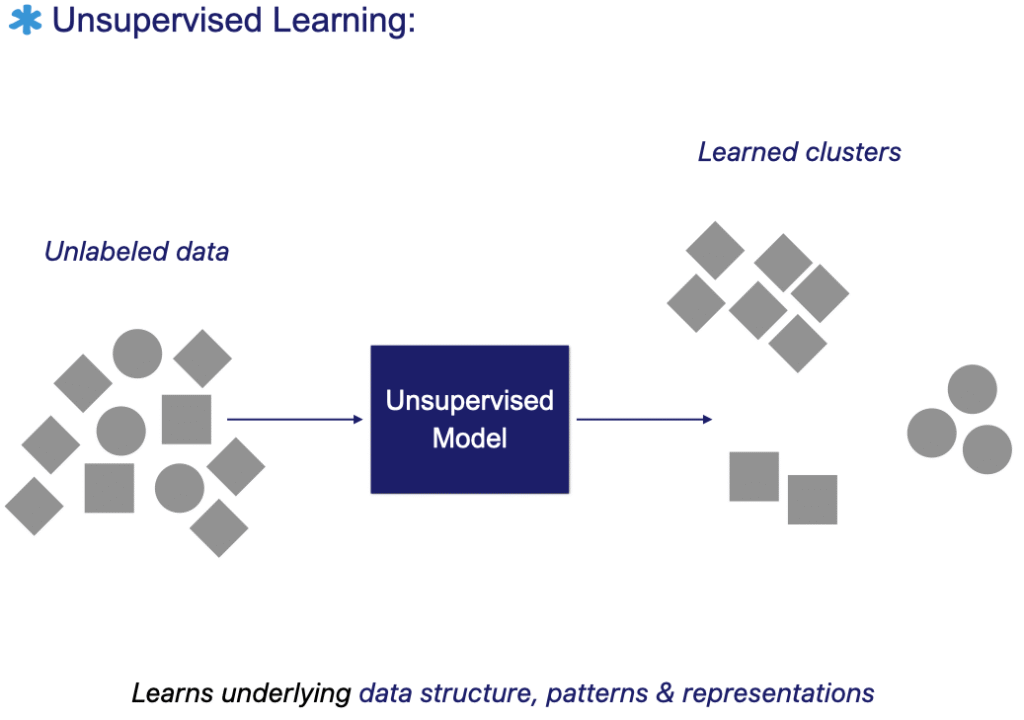
Unsupervised machine learning algorithms extract patterns and structure directly from raw data, without relying on predefined labels. The model learns to understand the underlying distribution of the data and can therefore group or cluster similar images.
Beyond clustering, unsupervised methods can learn compressed and meaningful representations that support data-driven decision-making. Their strength lies in their generality. These models often serve as a foundation for a wide range of downstream tasks.
Unsupervised learning is not limited to classification. It is also used for dimensionality reduction, compression, denoising, super-resolution, and even certain forms of autonomous decision-making. In practice, it is often advantageous to learn such representations before knowing the exact task they will serve.
The representations learned through unsupervised methods can later enhance the generalization and performance of supervised models trained on limited labeled data , as will be further illustrated in self-supervised learning.
Applications in medical imaging include:
- Segmentation: Prakram et al. reviewed unsupervised clustering-based methods for medical image segmentation, from segmenting lung nodules in CT scans and skin lesions in dermoscopy to identifying leukemia cells in microscope images and brain tumors in MRI.
- Registration: Liu et al. proposed ScaMorph, an unsupervised model for deformable image registration (DIR) that integrates convolutional neural networks with vision transformers. The model was evaluated across 3D tasks, including brain MRI and abdominal CT registration, and consistently outperformed previous methods.
- Anomaly detection: Baur et al. presented a comparative study on unsupervised anomaly detection (UAD) in brain MRI. Early work focused on clustering-based detection, while recent approaches combine generative models to directly detect and segment anomalies from imperfect reconstructions.
The figure below illustrates the unsupervised learning workflow: the model is trained directly on unlabeled data to discover patterns, clusters, or meaningful representations without predefined labels.
Self-supervised learning
Self-supervised learning (SSL), a branch of unsupervised learning, aims to extract meaningful and discriminative representations directly from unlabeled data, without requiring human annotations .
In SSL, the model learns by solving a pretext task that it generates from the data itself – for example, predicting a missing patch, estimating an image’s rotation, or determining whether two views originate from the same source.
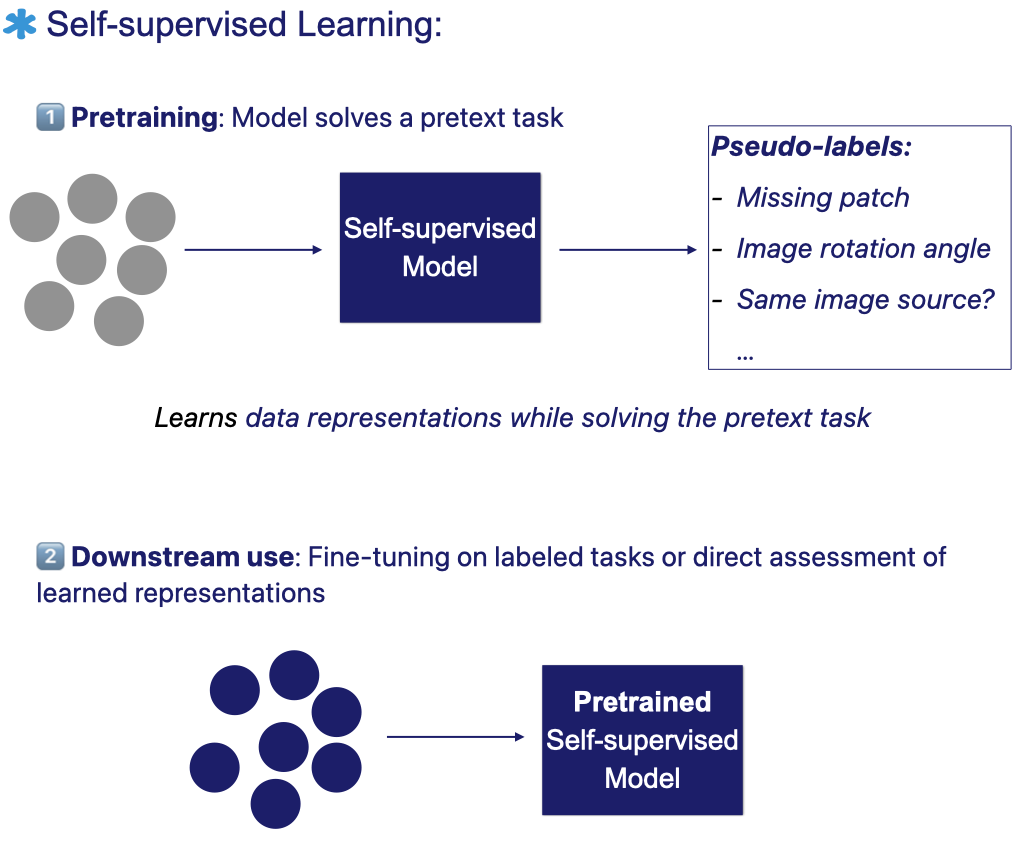
A typical SSL workflow involves two stages:
- Pretraining: the model solves a designed pretext task, where pseudo-labels are automatically generated from the data’s own structure and attributes.
- Fine-tuning or direct assessment: after pretraining, the model can either be fine-tuned on a labeled downstream task or directly evaluated to assess the quality of its learned representations .
Many studies have found that models perform better on downstream tasks when their feature extractors start from self-supervised pretrained weights rather than being trained from scratch with random initialization .
Pretraining generally helps models learn more efficiently, whether it’s used as an initial step with all labeled data or, more importantly, when labeled samples are scarce.
Applications in medical imaging:
A survey by VanBerlo et al. confirmed this trend, showing that self-supervised pretraining consistently improves downstream performance in radiography, CT, MRI, and ultrasound tasks.
The survey also highlights numerous clinical applications where SSL has proven beneficial – including breast cancer recognition in mammography, organ-at-risk segmentation in CT, brain and lesion segmentation in MRI, and fetal analysis in ultrasound imaging, among others.
The figure below illustrates the self-supervised learning workflow: the model first learns from unlabeled data through a pretext task with automatically generated pseudo-labels. It then transfers the learned representations to downstream tasks using labeled data.
Summary of learning paradigms
The following table summarizes how each learning paradigm handles labels and what it aims to achieve.
| Labels | Summary | |
|---|---|---|
| Semi-supervised | ✅ Few labeled dataset + many unlabeled ones | Combines labeled and unlabeled data in training |
| Unsupervised | ❌ None | Learns patterns or clusters directly from data |
| Self-supervised | 🧠 Generated automatically from pretext task | Learns from pseudo-labels derived from the data itself |
Conclusion
These techniques are transforming how we train AI in data-limited domains such as medical imaging. By learning from both labeled and unlabeled data, they enable more efficient use of existing resources, reduce dependence on costly manual annotations, and accelerate the translation of AI models into clinical practice.
In the upcoming articles of the Reducing Data Dependency series, I’ll explore each learning paradigm in greater depth, with practical examples, visual explanations, and insights from recent research.